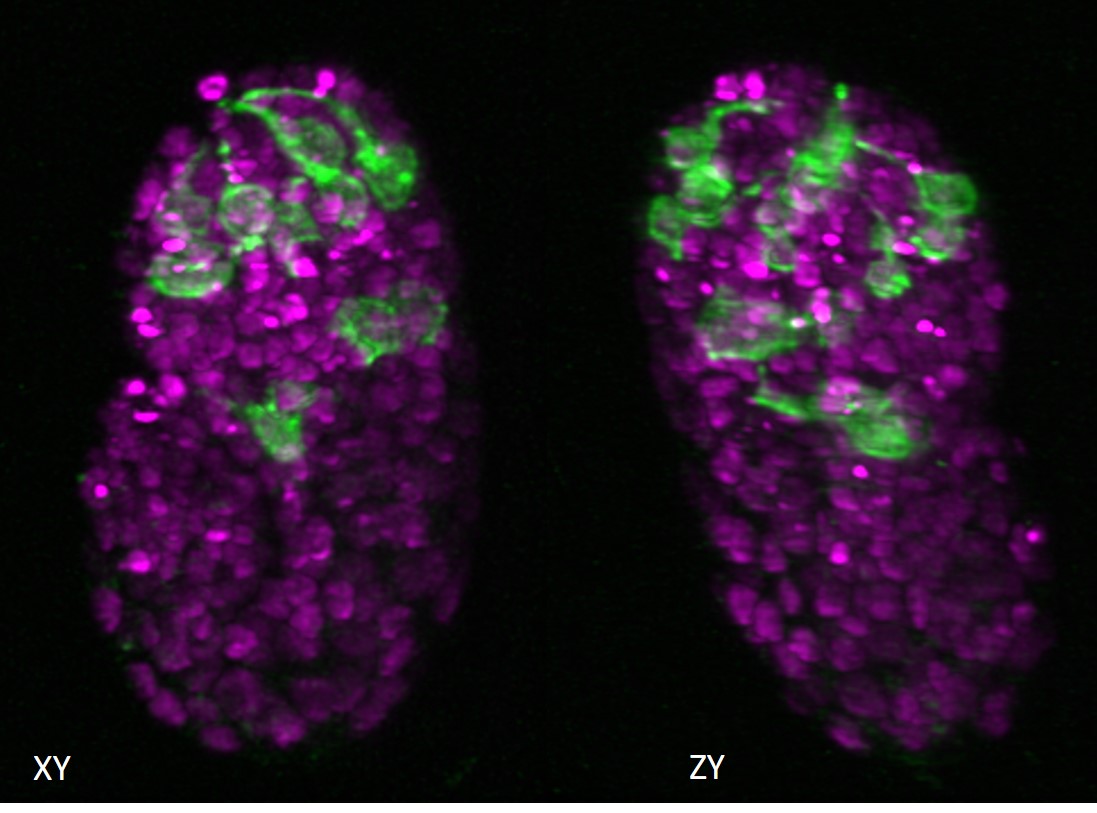
Scientists have developed new image processing techniques for microscopes that can reduce post-processing time by up to several thousand times. The researchers are from the National Institutes of Health and have collaborators from the University of Chicago and Zhejiang University, China.
in a article published in Nature BiotechnologyHari Shroff, Ph.D., head of the High Resolution Optical Imaging Laboratory at the National Institute of Biomedical Imaging and Bioengineering (NIBIB), describes new techniques that can significantly reduce the time needed to process the highly complex images created by the most advanced microscopes. These microscopes are often used to capture blood and brain cells moving through fish, visualize neuronal development in worm embryos, and locate individual organelles within entire organs.
As microscopes continue to improve and create higher-resolution images more quickly, researchers find that they have more data than they have time to process it. While the videos themselves can be captured in minutes, images can be terabytes in size and require weeks or, in some cases, months of processing time to make them usable.
One of the reasons it takes so long is that videos often capture tiny objects that are blurry under the microscope. This confusion can be reduced by a procedure called deconvolution, but this procedure requires a lot of computing power and time.
A second problem is that many of the microscopes used today take multiple views of the same organism or cell. Those images must be positioned correctly and then combined to create 3D images and videos. Creating high-resolution images from raw data requires a significant amount of computer processing. And so, while microscopes have been developed to provide researchers with increasingly complex, high-resolution images, computing power has limited the techniques that are practical for researchers to use, knowing that most of the data they collect will not be used.
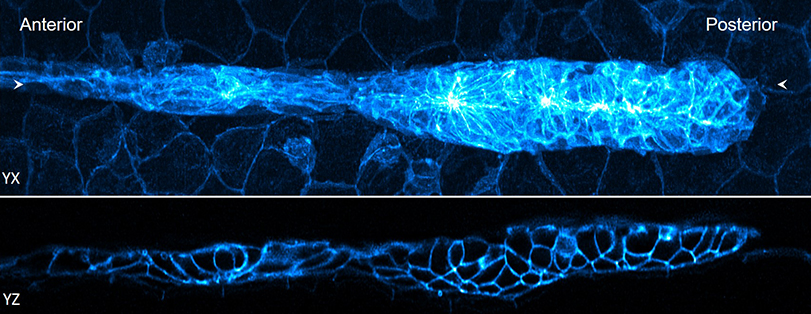
The first thing Shroff’s lab and his collaborators tried to do was modify the deconvolution algorithm that many researchers use, so that it would run faster. This approach was originally proposed for other areas of medical imaging, such as computed tomography (CT); however, this is the first time it has been successfully adapted for use with fluorescence microscopy. Fluorescence microscopy uses dyes to enhance sample contrast, allowing researchers to focus on specific parts of a sample and see how different elements interact with each other.
Second, they reduced the time needed to position and stitch multiple views of a sample. A key part of this advancement was based on a process called parallelization. It is an approach sometimes used in supercomputing where, instead of processing each individual function one after another, the work is divided into smaller tasks that can be analyzed simultaneously. It’s like asking thousands of people to solve a math problem simultaneously instead of asking one person to solve thousands of problems.
Finally, the researchers showed that they could further reduce the time it takes to process the data using a neural network, a type of artificial intelligence (AI). AI is increasingly being used to assist in image processing and diagnosis. In this case, Shroff and his team trained the neural network to produce cleaner, higher-resolution images much faster than would otherwise be possible.
“Acquiring modern imaging data is a bit like drinking from a fire hose,” Shroff said. “These methods help us obtain valuable biological information more quickly, which is essential given the enormous amount of data these microscopes can produce.”
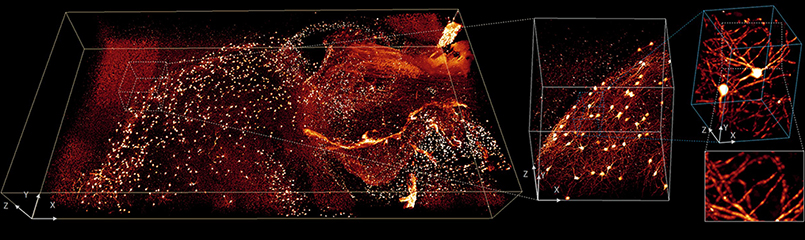
These advances expand the use of existing technology, including the ability to image thick samples that produce enormous amounts of image data when examined with fluorescence microscopes. Advances are also essential for the use of an increasing number of “computational microscopes” in which post-processing of unintelligible raw data is an essential step to produce the final high-resolution image. Shroff and his collaborators hope to help researchers with approaches they wouldn’t have thought to try, because of how laborious it would be to create meaningful images.
Scientists from the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Cancer Institute, Harvard University, the Sloan Kettering Institute, Yale University, the University of Maryland, and the University of Connecticut also contributed to this research.
Fast image deconvolution and multiview fusion for optical microscopy. Guo M, Li Y, Su Y, Lambert T, Nogare DD, Moyle MW, Duncan LH, Ikegami R, Santella A, Rey-Suarez I, Green D, Beiriger A, Chen J, Vishwasrao H, Ganesan S, Prince V, Waters JC, Annunziata CM, Hafner M, Mohler WA, Chitnis AB, Upadhyaya A, Usdin TB, Bao Z, Colón-Ramos D, La Riviere P, Liu H, Wu Y, Shroff H. Guo M, et al. Nat Biotechnology. June 29, 2020. doi: 10.1038/s41587-020-0560-x. PMID: 32601431.
###
NIBIB’s mission is to improve health by leading the development and accelerating the application of biomedical technologies. The Institute is committed to integrating the physical and engineering sciences with the biological sciences to advance basic research and healthcare. NIBIB supports research and development of emerging technologies within its internal laboratories and through grants, collaborations and training. More information is available on the NIBIB website: https://www.nibib.nih.gov.
The National Institutes of Health, the nation’s medical research agency, includes 27 institutes and centers and is a component of the U.S. Department of Health and Human Services. The NIH is the primary federal agency that conducts and supports basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures of common and rare diseases. For more information about NIH and its programs, visit https://www.nih.gov.