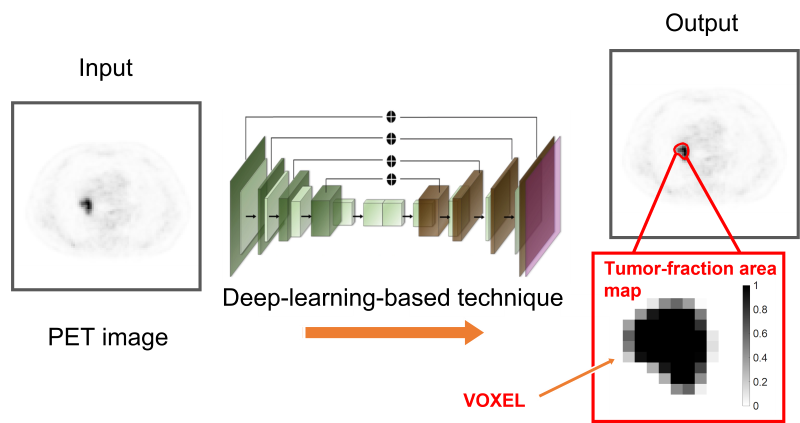
NIBIB-funded engineers are using deep learning to more accurately differentiate tumor from normal tissue in positron emission tomography (PET) images. Standard analysis of PET scans defines regions with abnormal radiotracer uptake as tumors. The team at Washington University in St. Louis has developed a technique that uses statistical analysis and deep learning to determine the extent of tumors at their margins.
PET images consist of what are known as voxels, which are three-dimensional pixels in space. Current methods count these voxels as tumor or normal.
“The key idea is that we can’t simply learn whether a voxel belongs to the tumor or not,” said team leader Abhinav Jha, Ph.D., assistant professor of biomedical engineering at the McKelvey School of Engineering. “The voxel can be partly tumorous and partly normal. The novelty is that we can estimate which part of the voxel is tumor.”
The research aims to provide more accurate information about the tumor to guide treatment decisions and improve patient care.
“It’s a quality of life issue for patients,” Jha said. “Helping answer those questions would be satisfying and rewarding.”
The work appears in the journal Physics in Medicine & Biology. [1]. The software is available for non-commercial purposes.
Financial support for this work was provided by the National Institute of Biomedical Imaging and Bioengineering R01 Award (R01-EB031051) and R21 Trailblazer Award (R21-EB024647), and a grant from NVIDIA. The University of Washington’s Center for High Performance Computing provided computational resources for the project. The center is partially funded by NIH grants 1S10RR022984-01A1 and 1S10OD018091-01.
[This is an update of the original post.]