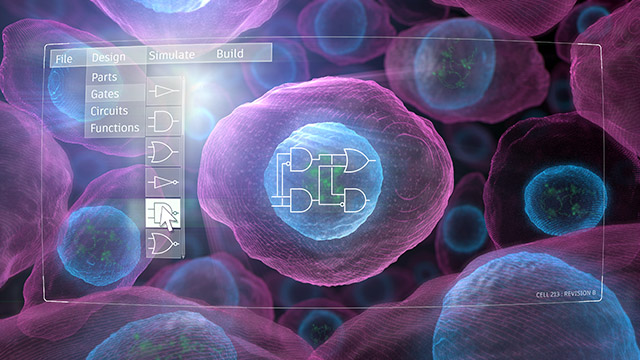
A change of instructions in a computer program tells the computer to execute a different command. Similarly, synthetic biologists are learning the rules of how to direct the activities of human cells.
“Cells are intricate machines that have evolved many interacting circuits: sets of genes that coordinate functions such as migration, metabolism, and cell division,” explains David Rampulla, PhD., director of the Science and Technology Discovery Division at the National Institute of Biomedical Imaging and Bioengineering. “Synthetic biologists aim to build genetic circuits that provide cells with new functions, which in the future could be used to control and treat diseases.”
However, a challenge in this field is that there are often many trial-and-error iterations required to create a circuit that works as intended. Now, NIBIB-funded synthetic biologists and computational modelers have teamed up to use computer simulations to avoid the laborious process of redesigning and retesting each genetic circuit.
One of the team members is engineer Josh Leonard, Ph.D., associate professor of chemical and biological engineering at Northwestern University’s McCormick School of Engineering. The computational modeler is Neda Bagheri, Ph.D., associate professor of biology and chemical engineering and Washington Research Foundation researcher at the University of Washington, Seattle. Together with their respective research teams, they are working to create genetic programs more quickly and reliably and to develop tools that help other researchers do the same.
“Engineering a cell comes down to building a piece of DNA, which encodes a set of genes whose products interact in a way we call a circuit or program. When that DNA is introduced into a cell, the cell is told to perform a desired function. Typically, it’s difficult to know if a circuit will work until we test it,” Leonard says. “This exceptional collaboration aims to create genetic programs that do what we want them to do the first time, every time, because computational models effectively take care of the trial and error up front.”

The team used custom simulations to analyze dozens of genetic circuits with different types of functions, such as turning genes on or off in response to various input signals. The most promising constructions were built and tested to see if they worked as intended. The research team was somewhat surprised to find that almost all of the circuits designed in this way closely matched the model’s predictions.
“In my experience, almost nothing works like that in science; nothing works the first time,” Leonard said. He explained that the usual process is to try many options, study the results, and debug to eventually identify a design that works.
Accelerated testing on computational models allowed the team to build and test operational circuits that performed increasingly complex tasks. Such tasks include programming the cell to evaluate signals from its environment, which could include proteins or other molecules that distinguish a healthy state from an imbalanced state, to decide whether to deliver a therapeutic payload.
“We are now at a point where we have a reliable set of tools and a formal design process for constructing gene regulatory functions,” said Joseph Muldoon, a recent PhD student and first author of the study. “It will next be exciting to see if these capabilities help address the unmet needs in biomedicine that motivated this work.”
The work is an important step toward efficiently designing a genetic “toolkit” that can be used by the broader biomedical engineering community to meet various application-specific needs. The team believes that new tools combined with computational models will allow bioengineers to create personalized cellular functions for applications ranging from fundamental research to new medical treatments.
The findings were reported in Science Advances. [1].
This work was supported by the National Institute of Biomedical Imaging and Bioengineering through award number 1R01EB026510, the National Institute of General Medical Sciences, the National Cancer Institute, the NUSeq Core of the Northwest Center for Genetic Medicine, an NU Chemistry of Life Processes Chicago Area Undergraduate Research Symposium award, and a NU Undergraduate Research Fellowship.