It may be surprising, but bacteria love to grow inside tumors. It’s the environment: Tumors are usually acidic, have low amounts of oxygen and are full of dead cells, which facilitates bacterial growth. Perhaps most importantly, the immune system cannot easily find and destroy microbes in the tumor core, giving bacteria a safe harbor to grow. This represents a therapeutic advantage: the bacteria can be used to attack cancerous tissues, penetrating deep into the tumor, where they can be engineered to release a therapeutic payload that kills cancer cells.
So why aren’t there more cancer treatments that incorporate engineered bacteria? One of the main issues is identifying which bacterial therapy would be most effective for a specific cancer type, which is a daunting and time-consuming task. Bacteria can determine how tumors grow and survive, and finding existing therapies that take advantage of these changes could lead to new treatment combinations for difficult-to-treat tumor types.
Now, researchers at Columbia University’s School of Engineering and Applied Sciences are developing a process to systematically evaluate how bacterial treatments might synergize with existing anticancer therapies. His studyrecently published in Nature scientific reportsused this process to evaluate treatment combinations in preclinical models of non-small cell lung cancer, which represent approximately 80% of all lung cancers.
“Lung cancers are a logical test case for bacterial treatments, as these tumors have been shown to harbor several different bacterial strains,” explained the study’s first author, Dhruba Deb, Ph.D., an associate research scientist at Columbia. “Although we chose to focus on non-small cell lung cancer in our study, our project could easily be adapted to focus on other types of cancer.”
Bacterial treatments have two important components: the type of bacteria used (such as Escherichia colifor example), and the payload that the bacteria are designed to produce, such as a toxin or other anti-cancer agent, which can destroy the cells. Both components can be modified to make bacterial treatments safer and more effective at killing cancer cells. The bacterial strain can be engineered to grow only in specific environments or to release its cargo only in the presence of a specific molecule, resulting in controlled release of the drug within tumors. The bacterial load can be designed or chosen so that it effectively kills specific types of cells.
In this study, the researchers used a bacterial strain that had been previously designed to release its cargo on demand, along with other modifications that made it less toxic. Regarding the therapeutic burden, the researchers focused on different types of bacterial toxins. The first step in their project was to identify which toxin had the best tumor-killing potential in non-small cell lung cancer. The researchers examined 10 bacterial toxins and found that theta toxin, a compound that punctures cell membranes, was the most effective at killing lung cancer cells. When researchers treated a mouse model of non-small cell lung cancer with bacteria engineered to release theta toxin, the lung tumors shrank markedly and there were no signs of damage to other organs.
Next, the researchers wanted to understand whether the bacterial treatment was affecting the underlying biology of lung tumors. “Cancer cells can evolve to better survive cancer treatments,” explained the study’s senior author, Tal Danino, Ph.D., an associate professor at Columbia. “If lung cancer cells become dependent on specific biochemical pathways for their survival after bacterial therapy, blocking these pathways with existing anticancer drugs could kill cancer cells more efficiently than either treatment approach alone,” Danino said.
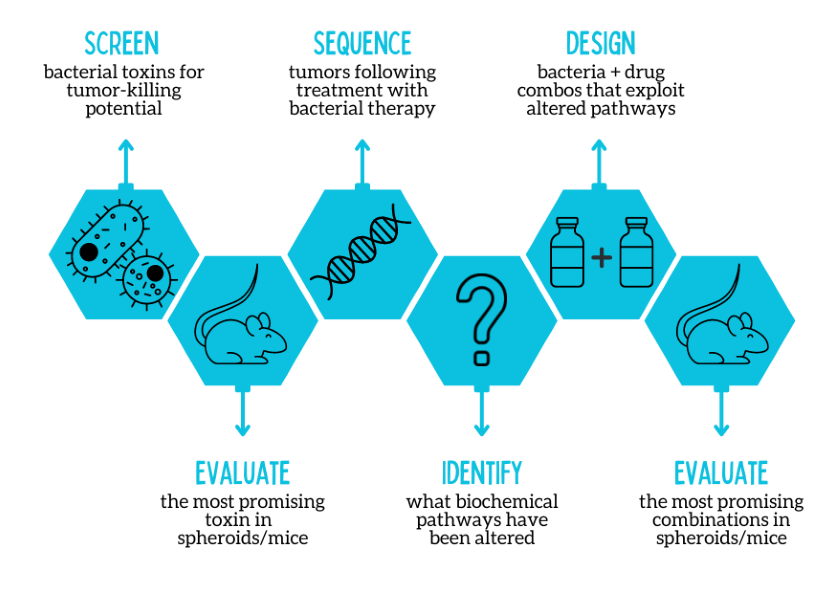
Using a next generation sequencing Using this approach, the researchers were able to identify which biochemical pathways were altered due to the bacterial therapy. It turns out that theta toxin affects specific pathways in lung cancer cells, such as those that control cell growth or DNA repair. By focusing on existing drugs that target these pathways, researchers could more efficiently screen for current anticancer drugs that could synergize with bacterial therapy.
After narrowing down the therapy combinations and evaluating them in tumor spheroids, the researchers evaluated the most promising combination in a mouse model of lung cancer. They found that mice treated with the combination therapy had the greatest reduction in tumor growth compared to mice treated with just one component of the treatment (either bacterial therapy or a drug targeting cell growth) alone.
“By incorporating next-generation sequencing methods and tumor spheroids, our pipeline could enable accelerated identification and evaluation of combinatorial treatments for lung cancer that incorporate bacterial therapy,” Deb said. “Globally, there are ~700 lung cancer therapies that are in various stages of preclinical and clinical trials. Our study opens up the possibility of choosing from this large repertoire of anticancer drugs to selectively combine with bacterial therapy for a precision medicine approach.”
“Engineered bacteria for cancer treatment represent an untapped area in drug development,” said David Rampulla, Ph.D., director of NIBIB’s Discovery Science & Technology division. “This study describes a potential way to accelerate the translation of bacterial therapies to the clinic, which could ultimately improve the effectiveness of existing cancer treatments.”
The researchers noted that humans are ~250 times more sensitive to bacteria than mice, and that further modifications to the bacterial strains will likely be necessary to keep this approach safe for human patients.
This study was supported by a NIBIB grant (R01EB029750).
This Science Highlight describes a basic research finding. Basic research increases our understanding of human behavior and biology, which is critical to promoting new and better ways to prevent, diagnose, and treat diseases. Science is an unpredictable and incremental process: each research advance builds on past discoveries, often in unexpected ways. Most clinical advances would not be possible without knowledge of fundamental basic research.
Study reference: Deb, D., Wu, Y., Coker, C. et al. Combination therapy design for engineered bacterial therapies in non-small cell lung cancer. science representative 12, 21551 (2022). https://doi.org/10.1038/s41598-022-26105-1